Internship proposals
Subject 1
Modeling and numerical simulation of neuron and neuronal networks morphogenesis

Left: dual culture chambers connected by simple diodes; top: experiment; middle: simulated neuronal network morphology; bottom: resulting connectome. Middle: experimental design of improved neurnal diodes (J. Courte et al. 2019). Right: simulated morphology.
Our group developed the DeNSE “Development of Neurons in Spatial Environments” simulator. As the name implies, this tool aims at enabling users to model the formation of neuronal morphologies in complex spatial environments, as well as the development of a neuronal network from the interactions of the separate neurons. This tool has recently been made publicly available online (https://github.com/SENeC-Initiative/DeNSE) but still needs extensive testing and parameterization in various situations.
Neuronal microfluidics devices are developed as research platforms for basic neurosciences and investigation of disorders of the central and peripheral nervous system.
This internship aims to explore neuronal network growth simulations with the DeNSE simulation. coupled with novel elastomer based microfluidics neural devices developed in the lab. The intern is expected to gain expertise with biophysical models of neuronal growth, biology and biophysics of neural cultures. The numerical platform will be used to implement new models and analyse a set of biological and biotechnological relevant biomedical situations in relation to experiments aiming to establish models on chips of myelinated peripheral nervous system.
This is a computational neurobiological and neurophysics project at the frontiers of neurobiology, physics, image analysis and programming.
Required skills:
Programming in Python (programming in C++ is a plus), Data analysis, Image analysis, Biophysical modelling, Interest for Neurosciences
Location:
Laboratory Matières et Systèmes Complexes, MSCmed group
Campus Universitaire des Saint Pères, 3rd floor, 45 rue des Saint Pères
Contact:
Prof. Samuel Bottani samuel.bottani@u-paris.fr
Stéphane Métens, stephane.metens@u-paris.fr
Hugo Salmon, hugo.salmon@u-paris.fr
Subject 2
Spatial bootstrap (quorum) percolation as a model for nucleation for neural activity
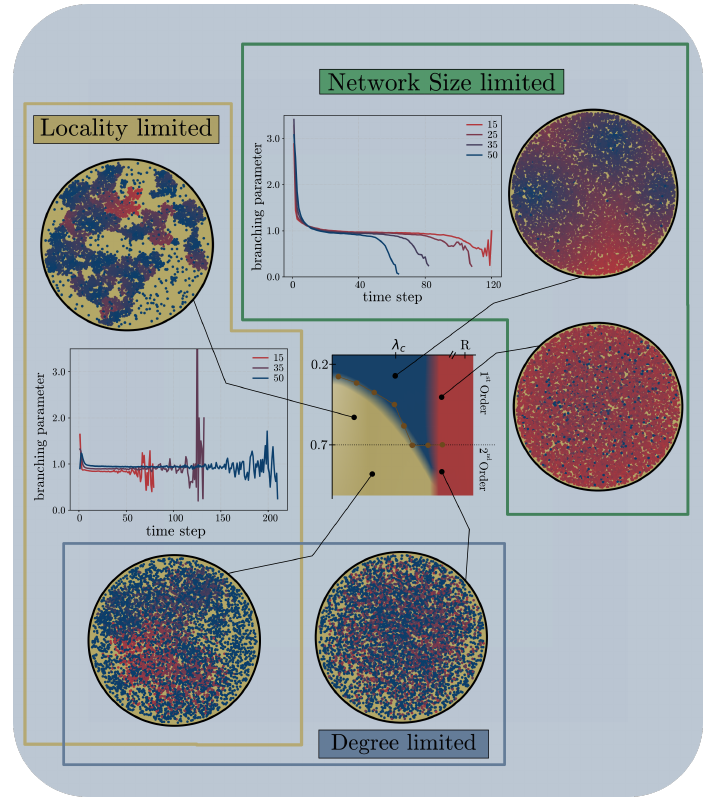
Phase diagram for different propagation dynamics. Blue region: localized nucleation dynamics. Yellow: fractured propagation dynamics. Red: non propagating dynamics.
Real, biological, neuronal networks of large number of cells form a specific tissue of active matter with complex emerging behaviors arising from the interplay of dynamical statistical and statistical systems.
Directed bootstrap percolation, also called quorum percolation, is an useful model of the response of neuronal population to external stimulations of increasing intensities. A purely topological version of this model (no space) has been used to indirectly investigate the unobservable ceonnectomics of the underlying network growing in a culture of biological neurons.
In this internship we propose to study the spatio-temporal dynamics of a space embedded quorum percolation model that displays different phase transitions in the nucleation and propagation dynamics of activation cascades.
Required skills : statistical physics,programing (pyhton), interest for neurosciences
Location:
Laboratory Matières et Systèmes Complexes, MSCmed group
Campus Universitaire des Saint Pères, 3rd floor, 45 rue des Saint Pères
Contact:
Prof. Samuel Bottani samuel.bottani@u-paris.fr
Stéphane Métens, stephane.metens@u-paris.fr